Michael Jahn, PhD
News
2025-01-09 | Genome sequence update of Streptococcus pyogenes published in MRA
We recently noticed problems to map next generation sequencing reads to certain regions of the Streptococcus genome, our pet organism for the study of pathogenic bacteria at the MPUSP. An extensive re-sequencing effort using short and long read methods revealed that the original assembly contained various errors, particularly for repetitive rRNA/tRNA operons. Our new assembly fixes these errors and adds two new tRNAs, becoming the current reference assembly for Streptococcus pyogenes. It’s available on NCBI datasets.
2024-10-08 | New paper on energy metabolism of Cupriavidus necator published in AEM
We published our second paper about C.necator using a barcoded transposon library to study its energy metabolism. This is not only a continuation of our previous work but also a systematic survey in the spirit of the landmark paper by R. Cramm, 2008 – in this extensive review, the author meticulously collected all known information about the energy metabolism of this interesting organism. Our work investigated the relative importance of all formate dehydrogenases, hydrogenases, and electron transport chain complexes experimentally, with several remarkable findings. This study was a fruitful collaboration between three groups that I am sincerely grateful for, Paul Hudsons lab, Oliver Lenz’ lab, and Emmanuelle Charpentier’s lab.
2023-07-26 | New CRISPRi library paper published in The Plant Cell
Very happy that our newest CRISPRi library paper was finally published in The Plant Cell (Oxford University Press). The paper takes a deep dive into the subtle tradeoffs between growth rate and growth robustness in photosynthetic bacteria. All fitness data are available online in my R Shiny App ShinyLib.
2023-06-09 | New CRISPRi library paper on BioRxiv
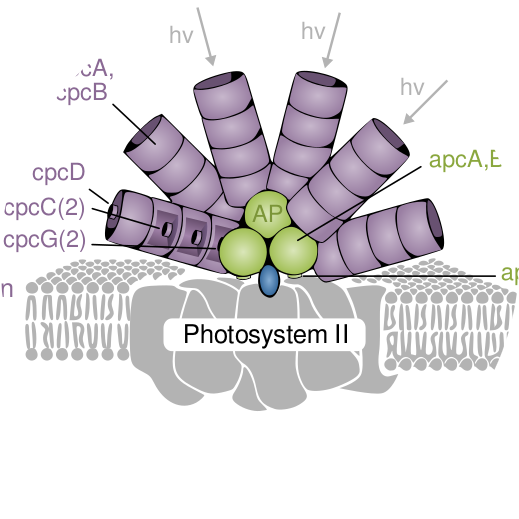
Together with my colleagues at Science for Life Lab, Stockholm, we have submitted a new manuscript with the second iteration of our CRISPR interference library in cyanobacteria. The paper is currently in revision but can already be accessed on BioRxiv.org. It’s filled to brim with new insights about fitness tradeoffs in photosynthetic bacteria, such as fitness cost for the highly expressed photosystem and light harvesting protein machines. Special thanks to Rui Miao who is the shared first author with me.
2023-06-06 | Joining the Editorial Board of Frontiers
I was recently invited to join the Editorial Board of the journal Frontiers in Bioengineering and Biotechnology. In the role of a review editor, I will occasionally review selected manuscripts for the journal and in return get public recognition for reviewing and editorial activities. This is more work for the me then for the journal, but it’s still an opportunity to be actively involved in the publishing process in my field. We will see how it goes.
2022-08-01 | New position at MPUSP, Berlin
New Job! After six exciting years as a Postdoc in Stockholm, I moved back to Germany together with my family. Living now in the greater Berlin area, I will start working at the Max Planck Unit for the Science of Pathogens (MPUSP) situated on the Charité campus in the heart of Berlin. I will work in the group of Emanuelle Charpentier, supporting different data-centered projects involving mass spectrometry, next generation sequencing, and many other things.
2021-11-11 | Finally published in eLife
Our new paper, “Protein allocation and utilization in the versatile chemolithoautotroph Cupriavidus necator”, was finally published in eLife!. It was a long journey, taking almost two and a half years to submission and another six months of review and improvements to finally publish it. We combined several layers of genome-wide screens: MS proteomics, gene essentiality from a transposon mutant library, and resource balance analysis modeling. Combining these layers allowed us to show that C. necator is a bacterium in the making. It has several conflicting layers of regulation to tune gene expression of its highly versatile but complicated carbon and energy metabolism. Our results show for example that cells “waste” protein resources for autotrophic lifestyle even when no hydrogen is available.
2021-03-24 | New preprint for Cupriavidus project
Our new paper about protein utilization in the versatile lithoautotroph Cupriavidus necator (aka Ralstonia eutropha) was just deposited on BioRxiv.org. Integrating data from proteomics, resource allocation modeling, and a transposon mutant library, we found some really interesting things. For example, of the three chromosomes that C. necator owns, one encodes 80% of the protein mass (the housekeeping chromosome). The other chromosomes encode specialized functions like CO2 fixation genes and were probably acquired later in evolution. The expression of these genes conflicts with overlapping functions of the ancestral chromosome.
2020-09-04 | Talk at cyano conference, Porto
In a few days starts the 11th European workshop on the biology of cyanobacteria. It was planned to take place in beautiful Porto, Portugal. I was very much looking forward to this as I haven’t been on a conference for a whole year. But unfortunately COVID-19 forced the conference organizers to turn it into a digital meeting. This is not as exciting as visiting a real conference (and a new city!) and meeting people in person, but I’m nevertheless looking forward to it. I will present my work on the cell economy of cyanobacteria. I recorded my talk (15 min) and uploaded it so you can watch it here.
2020-04-03 | CRISPRi library paper published
Finally, after 4 years of work with dozens of cultivations and millions of sequenced reads, our paper about a CRISPRi library in the model cyanobacterium Synechocystis was published in Nature Communications. What is this story about? Cyanobacteria are promising hosts for biotech production of (fine) chemicals. They are particularly interesting because they can grow solely on CO2 and sunlight. However, the productivities are low, the implemented pathways often instable, and the adaptation to varying amounts of light or CO2 not fully understood. Our group created a ‘library’ (a mixed pool) of thousands of mutants, each one equipped qith a CRISPR based gene repression system repressing one gene at a time. By cultivating this pool of mutants under low light, high light, and a lactate production condition, better adapted mutants were enriched over time, and worse adapted mutants depleted. By these means we were able to find a handful of genes improving growth, acid tolerance, or lactate productivity.
2019-11-08 | Awarded FORMAS grant
The Swedish funding agency FORMAS has awarded me with a grant to continue ongoing work with the fascinating litho-autotrophic bacterium Ralstonia eutropha a.k.a. Cupriavidus necator. As usual in our lab, we will study this bug and its metabolic capabilities from the systems biology perspective. Many exciting things are planned but the focus of the project is on resource allocation: What does a versatile soil bacterium spends its carbon and energy for? Which routes will different substrates take through the cellular metabolism? Under which conditions are enzymes saturated (under-abundance), or are not used at all (over-abundance)? We will try to answer these questions using highly controlled steady state cultivation, proteomics, metabolic modeling, and not yet determined genetic engineering. The project will officially start May 2020 and run for two years.
Short bio

- In 2022 I started as a bioinformatics researcher at the Max Planck Unit for the Science of Pathogens (MPUSP) in Berlin, Germany. The unit is headed by Nobel prize winner Emanuelle Charpentier and investigates a range of topics from RNA biology to CRISPR-Cas systems. The unit works with pathogenic bacteria such as Streptococcus pyogenes, the causative agent of scarlet fever in humans.
- From 2016 to 2022 I was a postdoctoral researcher in the group of Prof. Paul Hudson at Science for Life Lab at KTH, the Royal Institute of Technology in Stockholm, Sweden. My work focused on different projects evolving around microbial systems biology, mainly with photo- and lithoautotrophic model organisms, such as cyanobacteria or the ‘Knallgas bacterium’ Cupriavidus necator.
- From 2011 to 2015 I was doing my PhD at the Helmholtz Center for Environmental Research in Leipzig, Germany.
- My primary field of interest is the microbial world. I like to study bacteria at the interphase of experimental and computational biology. I am an enthusiastic user of R and python for statistics, metabolic modeling, and data visualization. I’m also advocating for openness and transparency in the life science. This website bundles information and resources of my scientific projects.
Quick links
Social media + platforms
Last updated: 2024-10-08
