Projects
This page gives an overview about ongoing (or finished) research projects. Every project contains a short description and links to bioinformatic resources and related publications.
- Cupriavidus necator energy metabolism
- Cupriavidus necator resource allocation
- New CRISPRi library in Synechocystis
- CRISPRi library in Synechocystis
- Synechocystis resource allocation
Cupriavidus necator energy metabolism
2021 - 2024
Published as The energy metabolism of Cupriavidus necator in different trophic conditions in Appl Environ Microbiol
Preprint is available at BioRxiv.org.
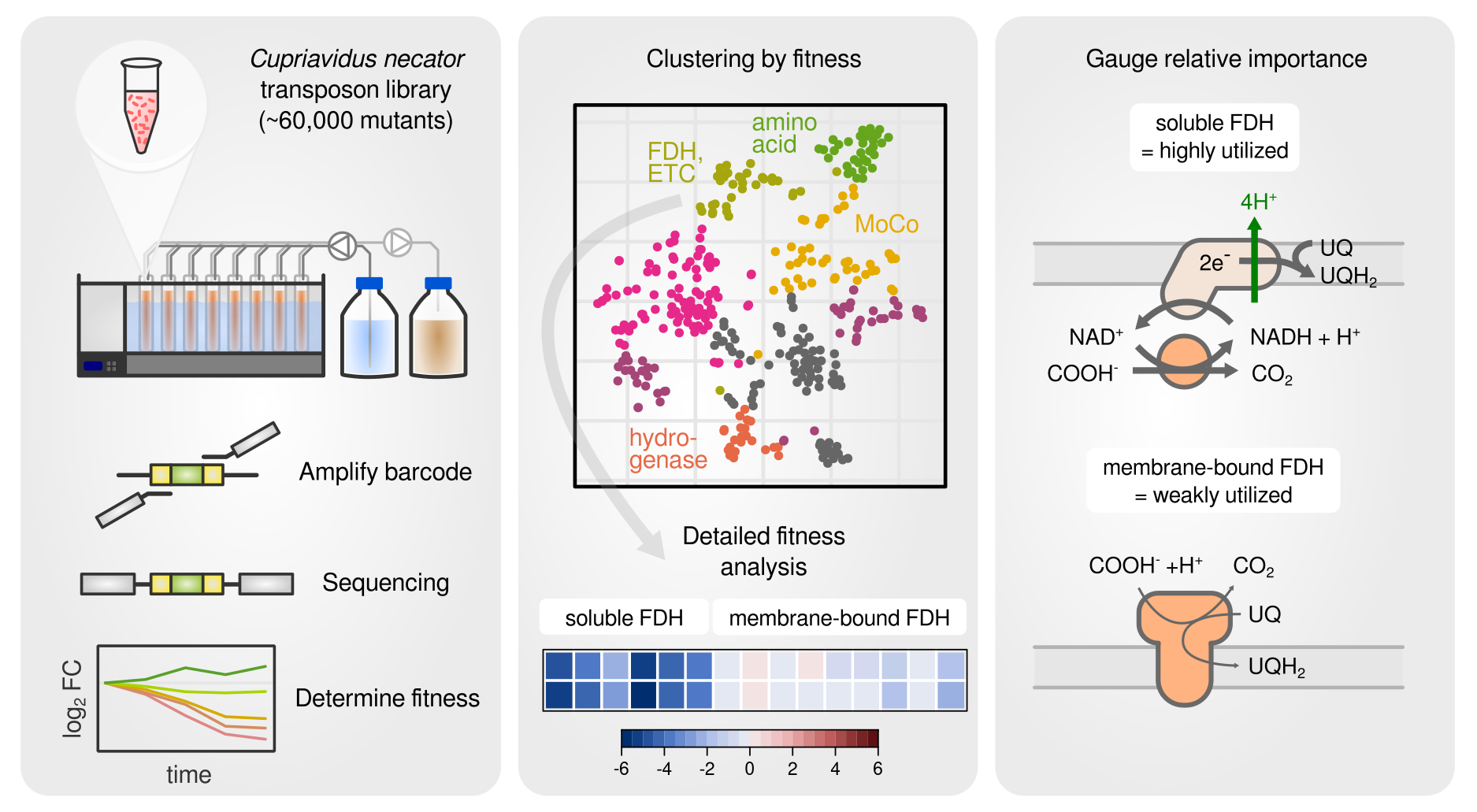
A barcoded transposon library was used to screen contribution of each gene to fitness.
In a continuation of the Cupriavidus necator resource allocation project, I am investigating the energy metabolism of this versatile chemolithoautotroph. A barcoded transposon knockout library is used to study the growth on different carbon and energy sources including the 1-carbon acid formic acid and hydrogen gas, which utilize Cupriavidus’ remarkable ability to fix CO2 and obtain energy from oxidation of small compunds such as formate or hydrogen. A particular focus was on gene fitness contribution for the molybdenum cofactor, the formate dehydrogenases, the various hydrogenases, and the electron transport chain. We found for example that C. necator utilizes both membrane bound and soluble hydrogenases for growth on hydrogen, but does only utilize the soluble formate dehydrogenase for growth on formate.
Resources
- ShinyLib app to browse fitness data
- ShinyProt app to browse proteomics data
- Giothub repo with analysis pipelines
Cupriavidus necator resource allocation
2019 - 2022
Published as Protein allocation and utilization in the versatile chemolithoautotroph Cupriavidus necator in eLife!
The preprint is available at BioRxiv.org.
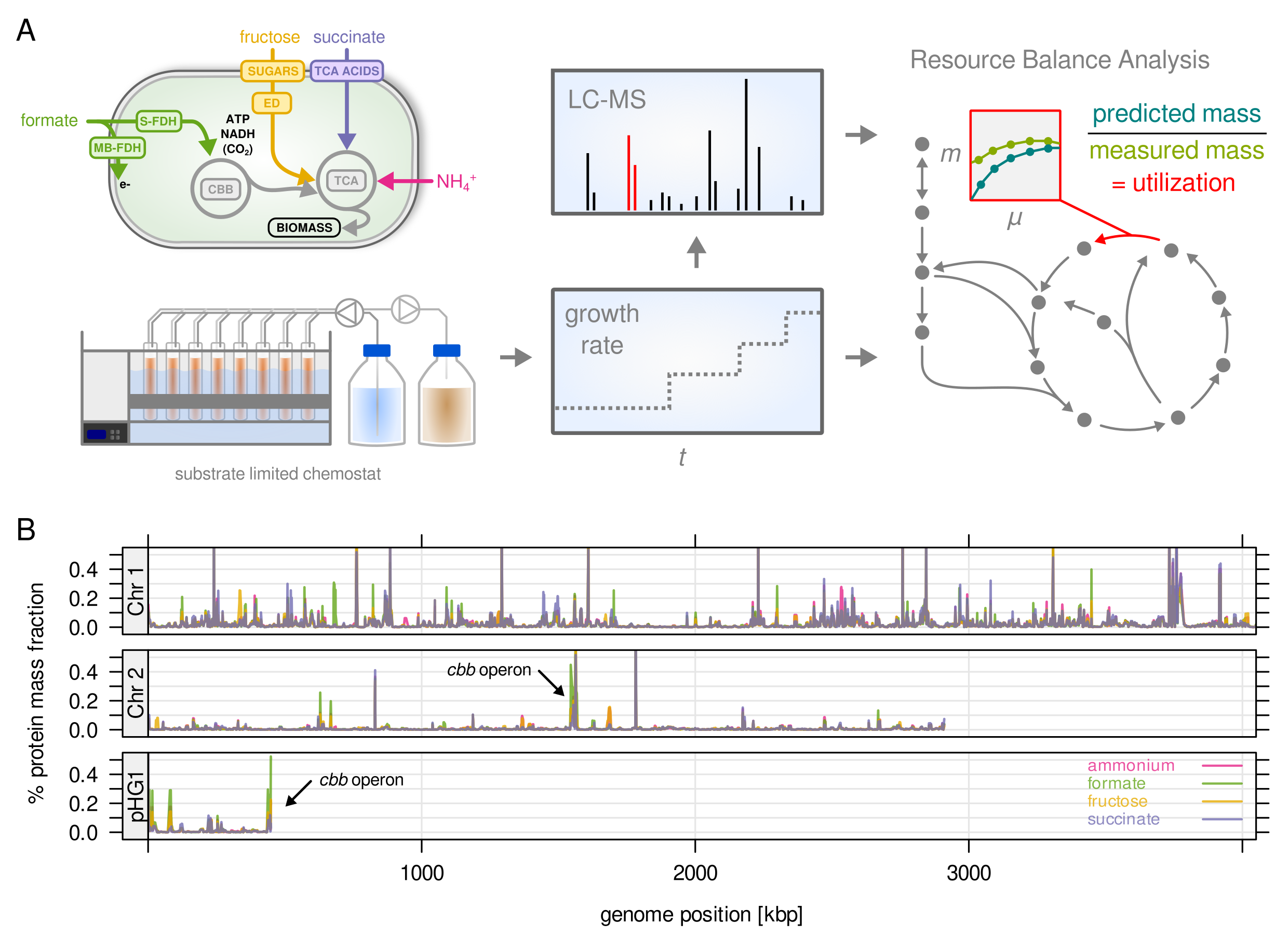
Study overview (A) and protein abundance mapped to the three chromosomes (B)
Ralstonia eutropha a.k.a. Cupriavidus necator is a highly interesting bacterium for its metabolic properties. It is a model organism both for the production of bioplastics (polyhydroxy butyrate, PHB) as well as for lithoautotrophic growth. This means it has the special ability to grow solely on CO2 as carbon source and hydrogen (H2) as energy source. These properties make it highly interesting for biotech applications, where we could picture a biorefinery taking CO2 and H2 as (waste) gas streams and feeding a culture of R. eutropha. The cells will then turn these substrates into biomass and product (such as PHB). However, R. eutropha is also a cryptic organism with a very large genome (6,600 genes), poorly understood metabolism and regulation, and very few genetic tools that are available.
In this project we want to address some of these shortcomings by analyzing growth under a variety of conditions. We will grow wild type and mutant strains with fixed growth rates (chemostat, turbidostat), gather information about gene expression using proteomics, and feed our data back into a mathematical model of the cell (see also Metabolic models for details). The focus of this project is to learn more about how a ‘large’ bacterium with many duplicated genes manages its gene expression resources. Bottlenecks and inefficiencies in gene expression will then be a starting point for the design of new engineered strains.
R notebooks for data analysis
- Ralstonia eutropha cultivations and physiological data
- Ralstonia eutropha processing of MS proteomics data
- Ralstonia eutropha model constraints
- Ralstonia eutropha RBA model, variability analysis
- Ralstonia eutropha enzyme abundance and utilization
- Ralstonia eutropha gene essentiality analysis (TnSeq)
- Ralstonia eutropha condition dependent fitness (BarSeq)
Web apps to browse the data
BioCircos maps showing the global gene expression profile of Ralstonia eutropha. These maps were made using a modified Biocircos R package.
- BioCircos map for Ralstonia eutropha, pHG1
- BioCircos map for Ralstonia eutropha, chromosome 1
- BioCircos map for Ralstonia eutropha, chromosome 2
New CRISPRi library in Synechocystis
2021 - 2023
Published as CRISPR interference screens reveal growth–robustness tradeoffs in Synechocystis sp. PCC 6803 across growth conditions in The Plant Cell
Preprint is available at BioRxiv.org.
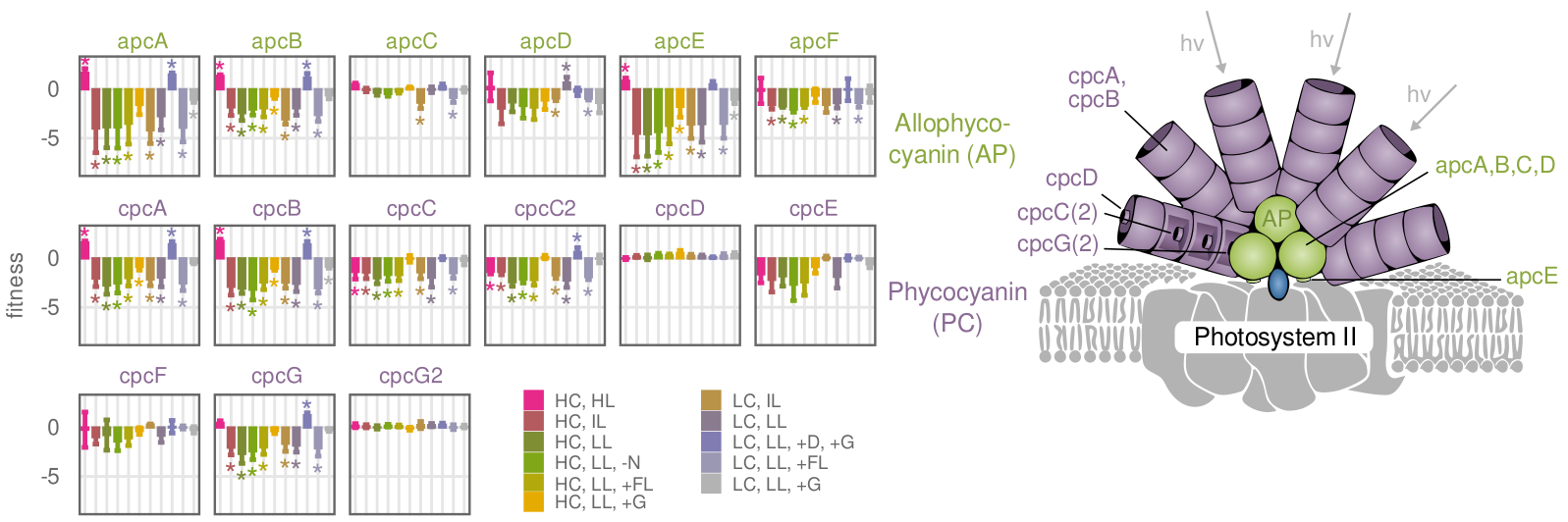
Light harvesting proteins show high fitness cost in intense light
This is the second iteration of a CRISPRi respression library for the cyanobacterium Synechocystis sp. PCC6803. The library is based on the inhibitory effect of the dCas9 gene and a corresponding short guide RNA (sgRNA) that conveys specificity to the enzyme. Repression of a gene will lead to fitness cost or benefit for the cell. By sequencing the pool of individual CRISPRi strains over time, these fitness effects become visible on a global scale. Besides some difficulties regarding repression efficiency of guide RNAs, this is the most powerful “functional genomics” screening method to date. In the corresponding paper, we addressed the problem of low guide efficiency by developing a machine learning model for efficiency prediction.
Resources
- Repository with all data analysis notebooks on github
- ShinyLib app to browse fitness data
- Nextflow pipeline for NGS screening data analysis
CRISPRi library in Synechocystis
2018 - 2020
Published as Pooled CRISPRi screening of the cyanobacterium Synechocystis sp PCC 6803 for enhanced industrial phenotypes in Nature Communications!
The preprint is available at BioRxiv.org.
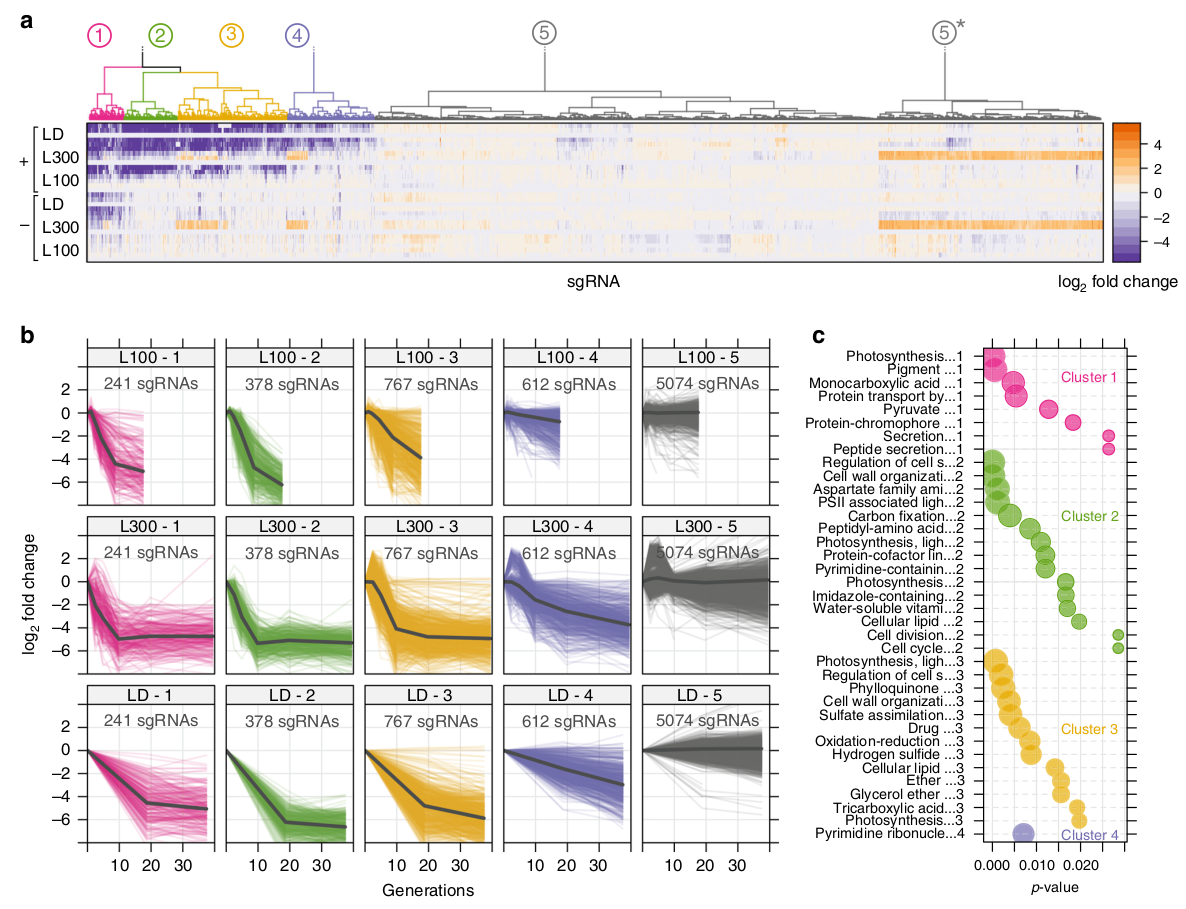
Example of depletion over time for single CRISPRi knockdown mutants
We designed a CRISPRi respression library for the cyanobacterium Synechocystis sp. PCC6803. The library is based on the inhibitory effect of the dCas9 gene and a corresponding short guide RNA (sgRNA) that conveys specificity to the enzyme. In each cell, the dCas9 enzyme will bind a unique sequence variant of the sgRNA. The dCas9-sgRNA complex then binds a region close to the promoter of the target gene and will repress transcription by physically blocking RNA polymerase. By using a pool of thousands of different sgRNAs, transcription of all genes of a bacterial strain can be repressed, one at a time.
R notebooks for data analysis
- Synechocystis CRISPRi library data processing
- Synechocystis CRISPRi library enrichment analysis
- Synechocystis CRISPRi library additional tests
Web app to browse the fitness data
Synechocystis resource allocation
2016 - 2018
Published as Growth of Cyanobacteria Is Constrained by the Abundance of Light and Carbon Assimilation Proteins in Cell Reports!
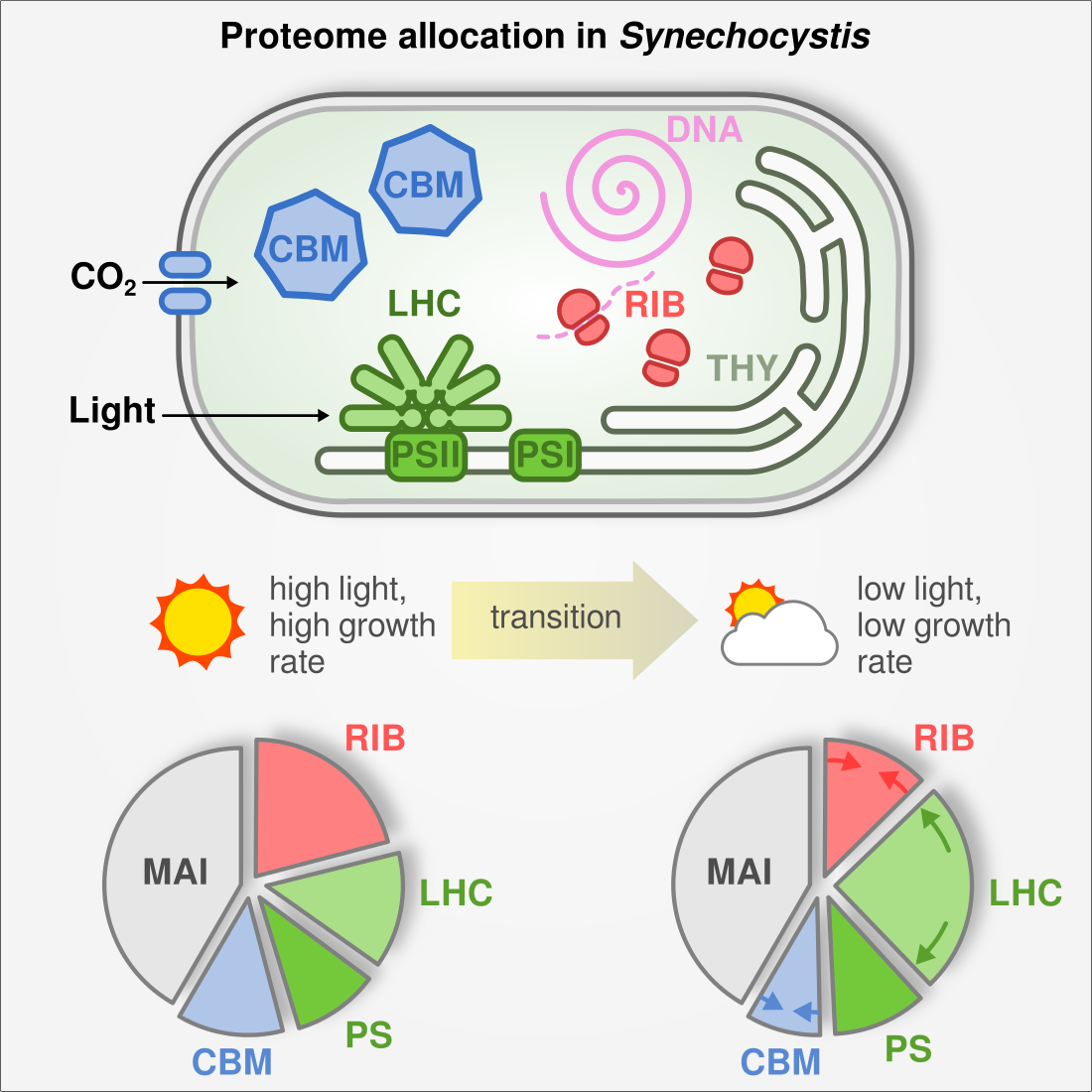
Cyanobacteria rearrange their proteome depending on light and CO2 availability
Cyanobacteria are promising hosts for microbial biotechnology as they convert inorganic CO2 and light into biomass and products. However, low biomass productivity has prevented wider industrial application. In this project we set out to investigate growth bottlenecks using gradual and systematic limitation of the energy and carbon source (light and CO2, respectively). We used shotgun proteomics to investigate protein allocation in the model cyanobacterium Synechocystis sp. PCC6803 as it adapts to limitation. We found that cells invest large amounts of resources into a small set of proteins. Of outstanding importance are proteins involved in carbon and light assimilation, as well as the translation machinery. When partitioning the proteome into a few functional sectors, we found that sector sizes changed linearly with growth rate. For example, the translation sector (ribosomes, chaperones) expands proportional to growth rate, confirming the ‘growth law’ in cyanobacteria. We fed our data into a coarse-grained cell economy model where protein costs are explicitly considered and growth rate is maximized. This demonstrated that cyanobacteria actively optimize protein resource allocation depending on environmental cues.
Resources
- Coarse grained cell economy models are described in Metabolic models
- Label-free proteomics pipelines for openMS/KNIME are deposited on github
Last updated: 2024-10-08
