Metabolic models
- Cupriavidus necator resource allocation model
- Cupriavidus necator genome scale model
- Synechocystis resource allocation model
Cupriavidus necator resource allocation model
Link to github repository with documentation

This resource balance analysis (RBA) model was generated using the RBApy package from Bulovic et al. (2019), using a genome scale metabolic model (GSM) originally developed by Park et al., 2011. The original genome scale model was refactored and many reactions were curated and harmonized to community standards as e.g. available through the Bigg model database. The genome scale model that serves as base for the RBA model is described below.
The RBA model is an extension of a standard GSM where the solution space is constrained by many additional parameters. The topology of the metabolic network and its gene and reaction annotations are taken from the GSM. In a next step, enzymes and cellular ‘machineries’ are built based on data retrieved from uniprot. The model simulates all cellular processes from DNA replication, to transcription, translation and protein folding. The protein pool is limited so that the optimization of gene expression towards a specific objective (e.g. growth) becomes dependent on environmental parameters but also rate limitations. Catalytic efficiency of enzymes links enzyme abundance to maximal rate so that flux through the metabolic netowrk can not become infinitely high.
Cupriavidus necator genome scale model
Link to github repository with documentation

Genome scale metabolic model for Ralstonia eutropha a.k.a. Cupriavidus necator H16. The model was previously published in Park et al., 2011. However the original model contained many errors or missing annotations and was therefore extensively manually curated. The full list of changes to the original model is documented on the models github repo. The original model, for example, showed flux through artificial energy generating cycles (see Fritzemeier et al., PLOS Comp Bio, 2017). After identification and removal of the following issues, no activity of such cycles was found anymore using FBA.
Synechocystis resource allocation model
Link to github repository with documentation
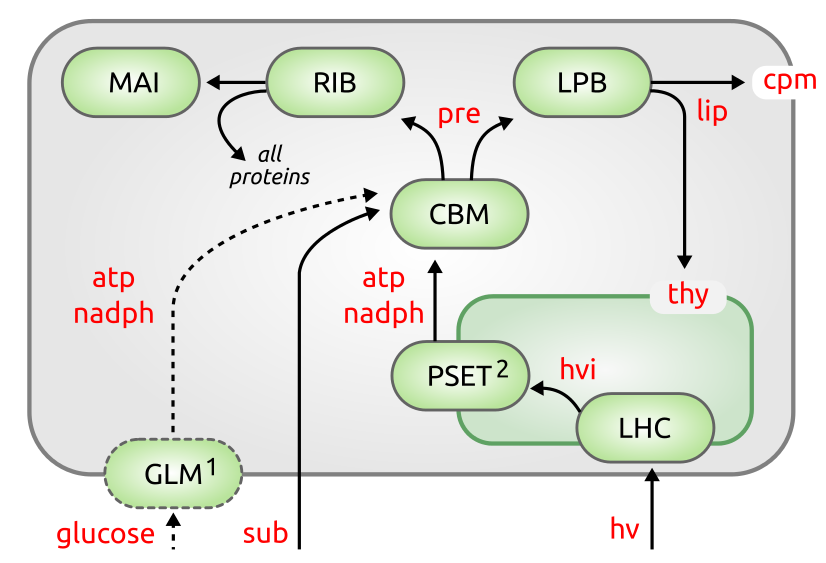
Simplified cellular economy model for the simulation of fundamental processes in photo-autotrophic bacteria. The purpose of this models is not to reflect cellular behavior in its entire complexity, but rather to reduce complexity to an amount that still allows drawing significant conclusions while keeping the number of components and reactions as small as possible. The cellular economy model bundles fundamental processes in ‘super-enzymes’, single catalytic units that serve as proxies for similar pathways. Such coarse-grain models are useful to illustrate metabolic tradeoffs under different conditions without getting lost in details. The models are mixed-integer non-linear optimization (MINLP) problems, based on previous work by Molenaar et al. (2009) and R. Burnap (2015).
Last updated: 2022-01-18
